Disclaimer
We assume no responsibility or liability for any loss or damage incurred as a result of any use of the information contained within or downloaded from this website.
If you have any problems, please let us know
WGE API
Regarding species
Each of the queries below requires a species parameter. This WGE instance accepts the following species:| species | Description |
|---|---|
| Grch38 | Human (GRCh38) |
| Mouse | Mouse (GRCm38) |
Crispr Search by Exon
Find CRISPRs for a given exonRequired fields: species, exon_id[]
Takes one or many ensembl exon ids:
https://wge.stemcell.sanger.ac.uk/api/crispr_search?species=Mouse&exon_id%5B%5D=ENSMUSE00000106755
Returns a json hash of exon ids -> crisprs:
{
"ENSMUSE00000106755":[
{
"ensembl_exon_id":"ENSMUSE00000106755",
"chr_start":35997419,
"off_target_summary_arr":[
"1",
"0",
"0",
"10",
"159"
],
"pam_right":0,
"species_id":2,
"exonic":1,
"chr_end":35997441,
"id":349738635,
"off_target_summary":"{0: 1, 1: 0, 2: 0, 3: 10, 4: 159}",
"genic":1,
"chr_name":"12",
"seq":"CCAGCCTTAAAGAAAGTGTTTGC"
},
...
]
}
Multiple exons can be specified like so:
https://wge.stemcell.sanger.ac.uk/api/crispr_search?exon_id%5B%5D=ENSMUSE00000106755&exon_id%5B%5D=ENSMUSE00000106761&species=Mouse
Crispr Search by Region
Find CRISPRs for a given region, returning GFF (for easy displaying in genoverse). A CRISPR will be included in the returned data if and only if its start point chr_start is within the region. I.e. if start <= chr_start <= end.Required fields: species_id, chr, start, end, assembly
IMPORTANT: This endpoint was originally created for internal use only, but was made public as it proved useful. Unfortunately, this means there are some peculiarities with regards to the interface.
- The assembly argument is in fact not used, except to set the text between parenthesis in the returned gff file. Thus any non-empty string can be used for this argument. However it IS necessary for the argument to be present, else you'll get an error.
-
To use a specific assembly you can set the species argument to that assembly, e.g. `species_id=Grch38`.
Valid arguments for the species_id parameter are currently:
- Human - which uses the GRCh37 assembly,
- Mouse - which uses GRCm38,
- Pig - which uses Sscrofa10.2,
- Grch38 - which is a more recent human assembly.
https://wge.stemcell.sanger.ac.uk/api/crisprs_in_region?start=35997423&end=35997496&species_id=Mouse&chr=12&assembly=GRCm38
Returns GFF:
##gff-version 3
##sequence-region lims2-region 35997423 35997496
# Crisprs for region Mouse(GRCm38) 12:35997423-35997496
12 WGE Crispr 35997423 35997445 . + . ID=C_349738636;Name=349738636;OT_Summary={0: 1, 1: 0, 2: 3, 3: 25, 4: 292}
12 WGE CDS 35997425 35997445 . + . ID=Cr_349738636;Parent=C_349738636;Name=349738636;color=#45A825
12 WGE CDS 35997423 35997425 . + . ID=PAM_349738636;Parent=C_349738636;Name=349738636;color=#1A8599
12 WGE Crispr 35997445 35997467 . + . ID=C_349738637;Name=349738637;OT_Summary={0: 1, 1: 0, 2: 6, 3: 259, 4: 1774}
12 WGE CDS 35997445 35997465 . + . ID=Cr_349738637;Parent=C_349738637;Name=349738637;color=#45A825
12 WGE CDS 35997465 35997467 . + . ID=PAM_349738637;Parent=C_349738637;Name=349738637;color=#1A8599
12 WGE Crispr 35997457 35997479 . + . ID=C_349738638;Name=349738638;OT_Summary={0: 1, 1: 0, 2: 0, 3: 13, 4: 162}
12 WGE CDS 35997459 35997479 . + . ID=Cr_349738638;Parent=C_349738638;Name=349738638;color=#45A825
12 WGE CDS 35997457 35997459 . + . ID=PAM_349738638;Parent=C_349738638;Name=349738638;color=#1A8599
12 WGE Crispr 35997472 35997494 . + . ID=C_349738639;Name=349738639;OT_Summary={0: 1, 1: 1, 2: 1, 3: 9, 4: 107}
12 WGE CDS 35997472 35997492 . + . ID=Cr_349738639;Parent=C_349738639;Name=349738639;color=#45A825
12 WGE CDS 35997492 35997494 . + . ID=PAM_349738639;Parent=C_349738639;Name=349738639;color=#1A8599
12 WGE Crispr 35997484 35997506 . + . ID=C_349738640;Name=349738640;OT_Summary={0: 1, 1: 0, 2: 1, 3: 5, 4: 85}
12 WGE CDS 35997486 35997506 . + . ID=Cr_349738640;Parent=C_349738640;Name=349738640;color=#45A825
12 WGE CDS 35997484 35997486 . + . ID=PAM_349738640;Parent=C_349738640;Name=349738640;color=#1A8599
12 WGE Crispr 35997490 35997512 . + . ID=C_349738641;Name=349738641;OT_Summary={0: 1, 1: 0, 2: 0, 3: 5, 4: 68}
12 WGE CDS 35997492 35997512 . + . ID=Cr_349738641;Parent=C_349738641;Name=349738641;color=#45A825
12 WGE CDS 35997490 35997492 . + . ID=PAM_349738641;Parent=C_349738641;Name=349738641;color=#1A8599
Crispr Pair Search by Exon
Find CRISPR pairs for a given exon, identical to the crispr_search but returns pairs instead of crisprs:https://wge.stemcell.sanger.ac.uk/api/crispr_search?exon_id%5B%5D=ENSMUSE00000106755&exon_id%5B%5D=ENSMUSE00000106761&species=Mouse
Returns JSON:
{
"ENSMUSE00000106755":[
{
"ensembl_exon_id":"ENSMUSE00000106755",
"right_crispr":{
"chr_start":35997445,
"off_target_summary_arr":[
"1",
"0",
"6",
"259",
"1774"
],
"pam_right":1,
"species_id":2,
"exonic":1,
"chr_end":35997467,
"id":"349738637",
"off_target_summary":"{0: 1, 1: 0, 2: 6, 3: 259, 4: 1774}",
"genic":1,
"chr_name":"12",
"seq":"ACATAAAGAAATCCAGAAATTGG"
},
"orientation":0,
"id":"349738635_349738637",
"left_crispr":{
"chr_start":35997419,
"off_target_summary_arr":[
"1",
"0",
"0",
"10",
"159"
],
"pam_right":0,
"species_id":2,
"exonic":1,
"chr_end":35997441,
"id":"349738635",
"off_target_summary":"{0: 1, 1: 0, 2: 0, 3: 10, 4: 159}",
"genic":1,
"chr_name":"12",
"seq":"CCAGCCTTAAAGAAAGTGTTTGC"
},
"db_data":null,
"spacer":3
},
...
]
}
Off-Targets for Crisprs
Fetch off-target summaries and list of off-target crispr IDs for 1 or more crisprs (up to max of 100).Required: species, id
https://wge.stemcell.sanger.ac.uk/api/crispr_off_targets?id=1106710989&id=1106710985&species=Grch38
Returns an object mapping crispr ID to its off-target summary and off-target list
{
"1106710985": {
"off_targets": [
904032520,
904764939,
...
1197488029,
1199013883
],
"off_target_summary": "{0: 1, 1: 0, 2: 0, 3: 2, 4: 66}",
"id": 1106710985
},
"1106710989": {
"off_targets": [
902582231,
906234136,
...
1188165849,
1201450411
],
"off_target_summary": "{0: 1, 1: 0, 2: 0, 3: 4, 4: 49}",
"id": 1106710989
}
}
Off-Targets for Crispr Pairs
Fetch an off-target summary and list of off-target pair IDs for a crispr pairRequired: species, left_id, right_id
where left_id is the CRISPR ID of the left crispr and right_id is the CRISPR ID of the right CRISPR.
Note: this method can be slow (10-20 seconds) if the off-targets for this pair have not been pre-computed
https://wge.stemcell.sanger.ac.uk/api/crispr_pair_off_targets?left_id=322289790&right_id=322289791&species=Mouse
Returns an json string mapping the crispr pair ID to its off-target summary and list of off-target IDs.
{
"1106711016_1106711017": {
"off_targets": [
"1106711016_1106711017"
],
"off_target_summary": "{\"closest\":\"None\",\"total_pairs\":1,\"max_distance\":1000}",
"id": "1106711016_1106711017"
}
}
Find Crispr ID for Sequence
Find a CRISPR ID for a given gRNARequired fields: seq, species, pam_right
pam_right can be set to the following values:
- 0 - only find crisprs on the global negative strand
- 1 - only find crisprs on the global positive strand
- 2 - search in both orientations
https://wge.stemcell.sanger.ac.uk/api/search_by_seq?species=Mouse&pam_right=2&seq=GTCCCCAGAATTGTGTTTGT
Returns a list of IDs that matched:
[349738765]Or, with get_db_data set to 1 a list of CRISPRs:
[
{
"chr_start":35998625,
"off_target_summary_arr":[
"1",
"0",
"0",
"10",
"155"
],
"pam_right":1,
"species_id":2,
"exonic":1,
"chr_end":35998647,
"id":349738765,
"off_target_summary":"{0: 1, 1: 0, 2: 0, 3: 10, 4: 155}",
"genic":1,
"chr_name":"12",
"seq":"GTCCCCAGAATTGTGTTTGTAGG"
}
]
Find Off-Targets for Sequence
Fetch off-target summary and list of off-target crispr IDs for any 20bp sequence.Required fields: seq, species, pam_right
pam_right must be set to 'true' or 'false'
https://wge.stemcell.sanger.ac.uk/api/off_targets_by_seq?seq=TTAATTGGTCAGCCTAACTC&species=mouse&pam_right=false
Returns off-target summary and off-target list. If a CRISPR site is found in the genome that exactly matches the search sequence then the ID of this is given. If there are multiple exact matches then the ID is that of the first match. If no exact match is found in the genome the ID returned will be 0.
{
"off_targets": [
302072111,
310736349,
310901261,
320182042,
456345367,
...
],
"off_target_summary": "{0: 1, 1: 0, 2: 0, 3: 6, 4: 48}",
"id": 456345367
}
Find Crispr Sequence by ID
Fetch Crispr sequence for 1 or more crispr IDs.Required: species, id
https://wge.stemcell.sanger.ac.uk/api/crispr_seq_by_id?id=1106710999&id=1106711006&species=Grch38
Returns an JSON containing the crispr sequence:
{
"1106711006":
{
"seq":"GCCATTAAATGAGGAAACAGTGG"
},
"1106710999":
{
"seq":"CCTATTGCATATTTCTTCATGTG"
}
}
Find Crispr by ID
Fetch Crispr information and an off-target summary for 1 or more crispr IDs.Required: species, id
https://wge.stemcell.sanger.ac.uk/api/crispr_by_id?species=Grch38&id=1106710999&id=1106711006
Returns an JSON containing the crispr:
{
"1106711006":
{
"chr_start":32332849,
"pam_right":1,
"species_id":4,
"exonic":1,
"chr_end":32332871,
"id":1106711006,
"off_target_summary":"{0: 1, 1: 0, 2: 1, 3: 30, 4: 301}",
"genic":1,
"chr_name":"13",
"seq":"GCCATTAAATGAGGAAACAGTGG"
},
"1106710999":
{
"chr_start":32332714,
"pam_right":0,
"species_id":4,
"exonic":1,
"chr_end":32332736,
"id":1106710999,
"off_target_summary":"{0: 1, 1: 0, 2: 3, 3: 46, 4: 563}",
"genic":1,
"chr_name":"13",
"seq":"CCTATTGCATATTTCTTCATGTG"
}
}
Find Marker Symbols for Search Term
Find all marker symbols matching your search, with wildcards either side of your value:Required fields: name, species
https://wge.stemcell.sanger.ac.uk/api/gene_search?species=Mouse&name=cb
Returns a list of marker symbols:
["Abcb10", "Abcb11", "Abcb1a", "Abcb1b", "Abcb4", "Abcb5", "Abcb6", "Abcb7", "Abcb8", "Abcb9", "Acacb", "Acbd3", "Shcbp1", "Shcbp1l", "Smarcb1", "Sncb", "Tbcb", "Ube2cbp"]
Find Exons for Marker Symbol
Find all exons for a marker sybmol. This is used by the find_crisprs table.Required: marker_symbol, species
https://wge.stemcell.sanger.ac.uk/api/exon_search?species=Mouse&marker_symbol=Cbx1
Returns an object containing the transcript id and a list of exons:
{
"exons":[
{
"len":195,
"exon_id":"ENSMUSE00000761328",
"id":"736362",
"rank":1
},
...
{
"len":457,
"exon_id":"ENSMUSE00000585969",
"id":"736364",
"rank":6
}
],
"transcript":"ENSMUST00000093943"
}
Off-Targets for Crisprs (DEPRECATED)
This method has now been replaced by crispr_off_targets which returns off-target information for all IDs queried
Calculate off-targets for individual CRISPRs, persisting into WGE.Required: species, ids[]
ids[] is a list of CRISPR IDs that do not have off-target data. Any IDs provided that already have off-target data will be ignored
https://wge.stemcell.sanger.ac.uk/api/individual_off_target_search?species=Mouse&ids%5B%5D=322289792
Returns an object in JSON mapping CRISPR id -> off target data ONLY for CRISPR IDs that were updated. If there were no CRISPRs needing off-targets the returned object will be empty. A maximum of 100 CRISPR IDs will be processed.
{
"322289792":{
"off_targets":[
"300894653",
"300947249",
"301353461",
"301376066",
...
"573722530",
"573805949",
"576503162",
"576681220"
],
"off_target_summary":"{0: 1, 1: 0, 2: 1, 3: 35, 4: 413}",
"id":322289792
}
}
Off-Targets for Crispr Pairs (DEPRECATED)
This method has been replaced by crispr_pair_off_targets which returns the list of off-targets identified for the crispr pair
Calculate off-targets for a single CRISPR pair, persisting into WGE.
Required: species, left_id, right_id
where left_id is the CRISPR ID of the left crispr and right_id is the CRISPR ID of the right CRISPR.
Note: this method can be slow (10-20 seconds per pair)
https://wge.stemcell.sanger.ac.uk/api/pair_off_target_search?right_id=322289791&species=Mouse&left_id=322289790
Returns an json string with two fields:
{success: 1, pair_status: 5}
WGE Components
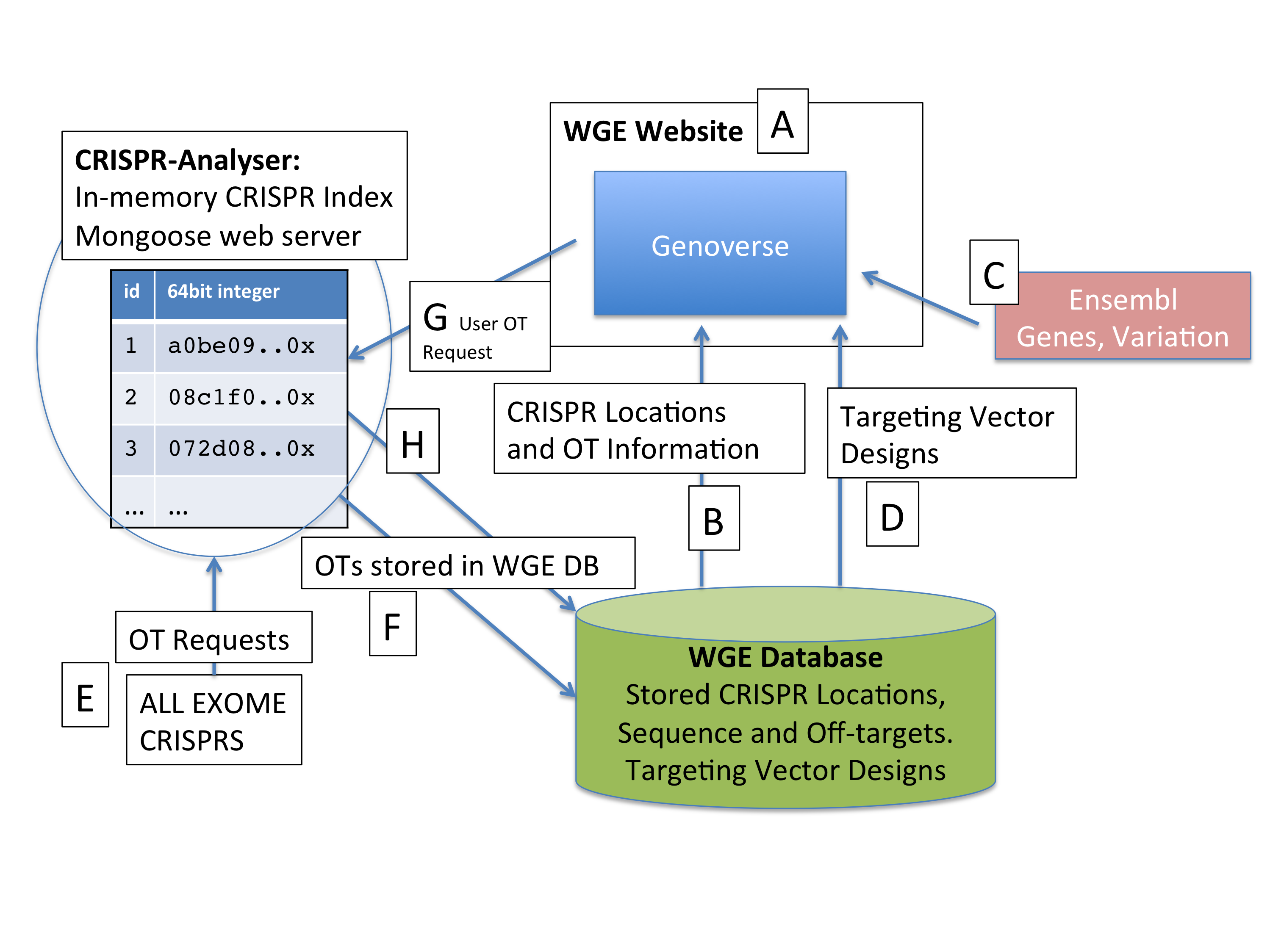
(A) The WGE website (with the genoverse genome browser) presents pre-computed CRISPR and off-target data from the WGE Database (B) as well as Ensembl gene structure and variation data (C), and user-generated targeting vector designs (D). CRISPR location data for the whole genome is pre-computed in WGE. Off-target data is precomputed for all CRISPRS in the exome (E) and stored in WGE (F). Off-target data which has not been pre-computed can be requested from the website (G) and is stored in WGE (H) and displayed as usual (B).
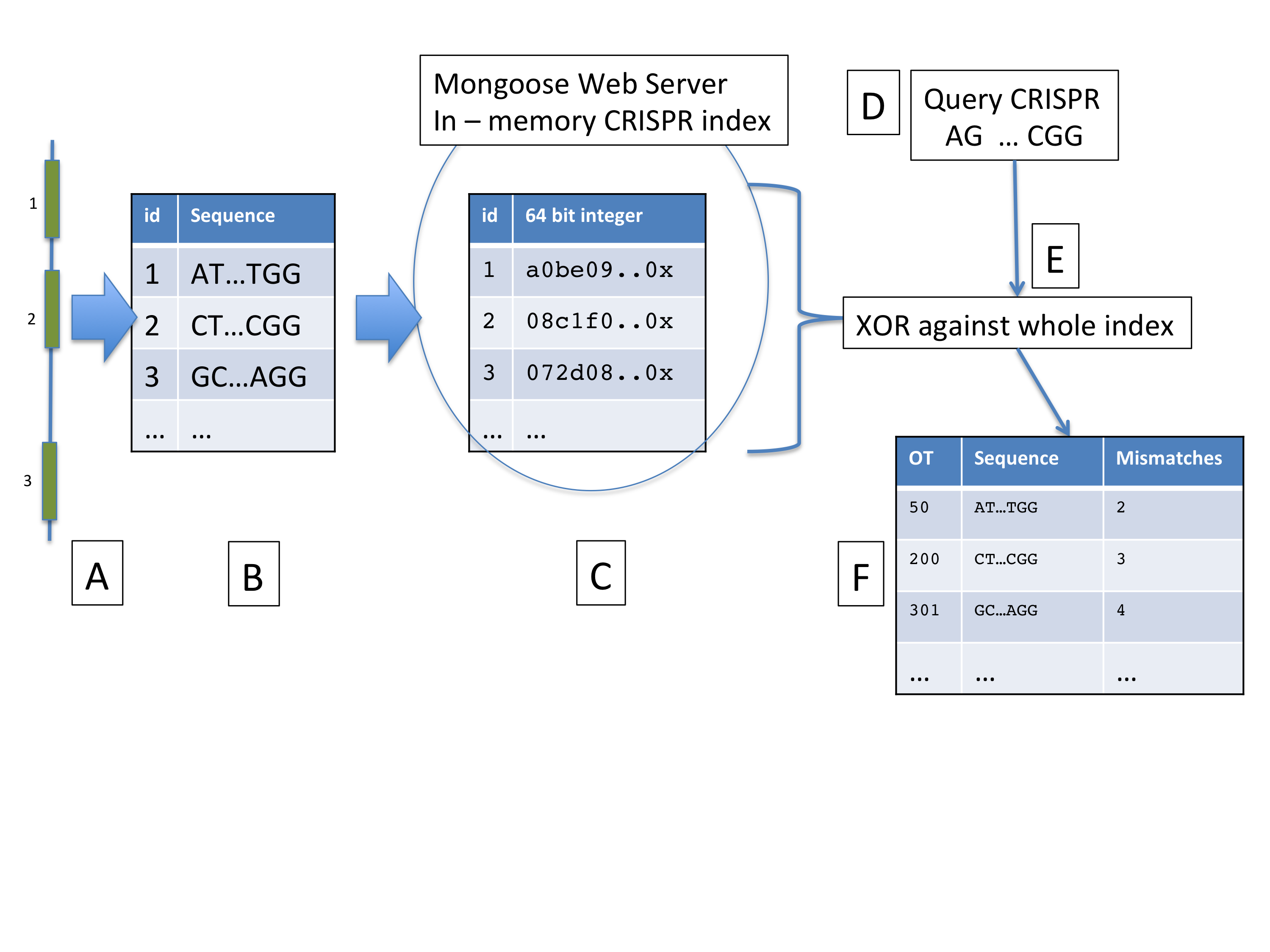
CRISPR-Analyser
To calculate off targets for CRISPRs we use the CRISPR-Analyser package, hosted here: CRISPR-Analyser on github
The CRISPR-Analyser works as follows: (A) Any Genome is scanned for all possible CRISPR sites (1,2,3â¦), and the resulting sequences (B) are stored as a CSV file. (C) Each CRISPR sequence in the file is converted to a 64 bit integer and the resulting index kept in-memory is a Mongoose server. All targets for a specific query-CRISPR string (D) are matched against every possible genomic crispr site using a rapid XOR (E) and the resulting possible Off-target sites returned as a list (F), along with the number of mismatches to the possible off-target site.
Source code
Our web application is written in Perl using the Catalyst framework. The application runs on a PostgreSQL database.
The code can be seen in our github repository